TPWshiny: An Interactive R/Shiny App to Explore Cell Line Transcriptional Responses to Anti-cancer Drugs
Published in Bioinformatics, 2021
Recommended citation: Zhang, P., Palmisano, A., Kumar, R., Li, M. C., Doroshow, J. H., & Zhao, Y. (2021). TPWshiny: an interactive R/Shiny app to explore cell line transcriptional responses to anti-cancer drugs. Bioinformatics. https://doi.org/10.1093/bioinformatics/btab619
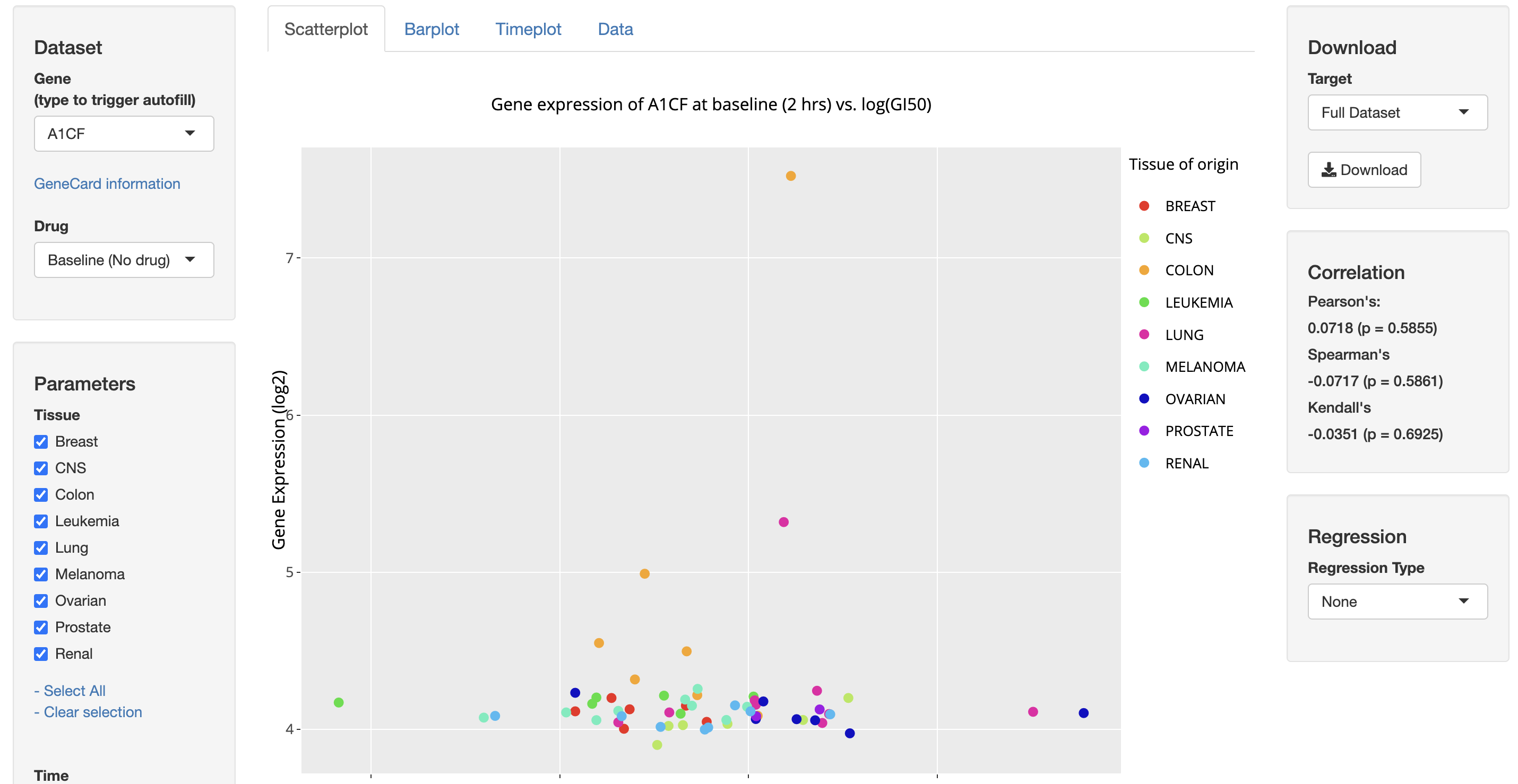
From August of 2019 to August of 2021, I collaborated with a team in the Biometrics Research Program at the National Cancer Institute led by Dr. Yingdong Zhao. We developed TPWshiny, an R Shiny application for visualizing data from NCI-60 human cell lines.
For cancer researchers, the NCI’s Transcriptional Pharmacodynamics Workbench (TPW) is an important dataset that documents the responses of cancer cell lines to common anticancer agents. It can yield insights about the effectivess and weaknesses of drugs. The application is live on the NCI’s website and is free to use by anyone. It supports analysis of single genes, geneset patterns, genes over time, and sensitivity/resistance.